Welcome to our Interactive Virome Analysis Tool!
This application was designed in R Shiny to explore interactively the results of a project involving the analysis of viromes in 417 public RNA-Seq datasets of invertebrates.
Explore!
Methods & code availability
Methods used for the taxonomic classification pipeline are explained in the original publication. Code and data for this app and the procedures described in the article are available at https://git.csic.es/sfelenalab/invertebrates-virome. .
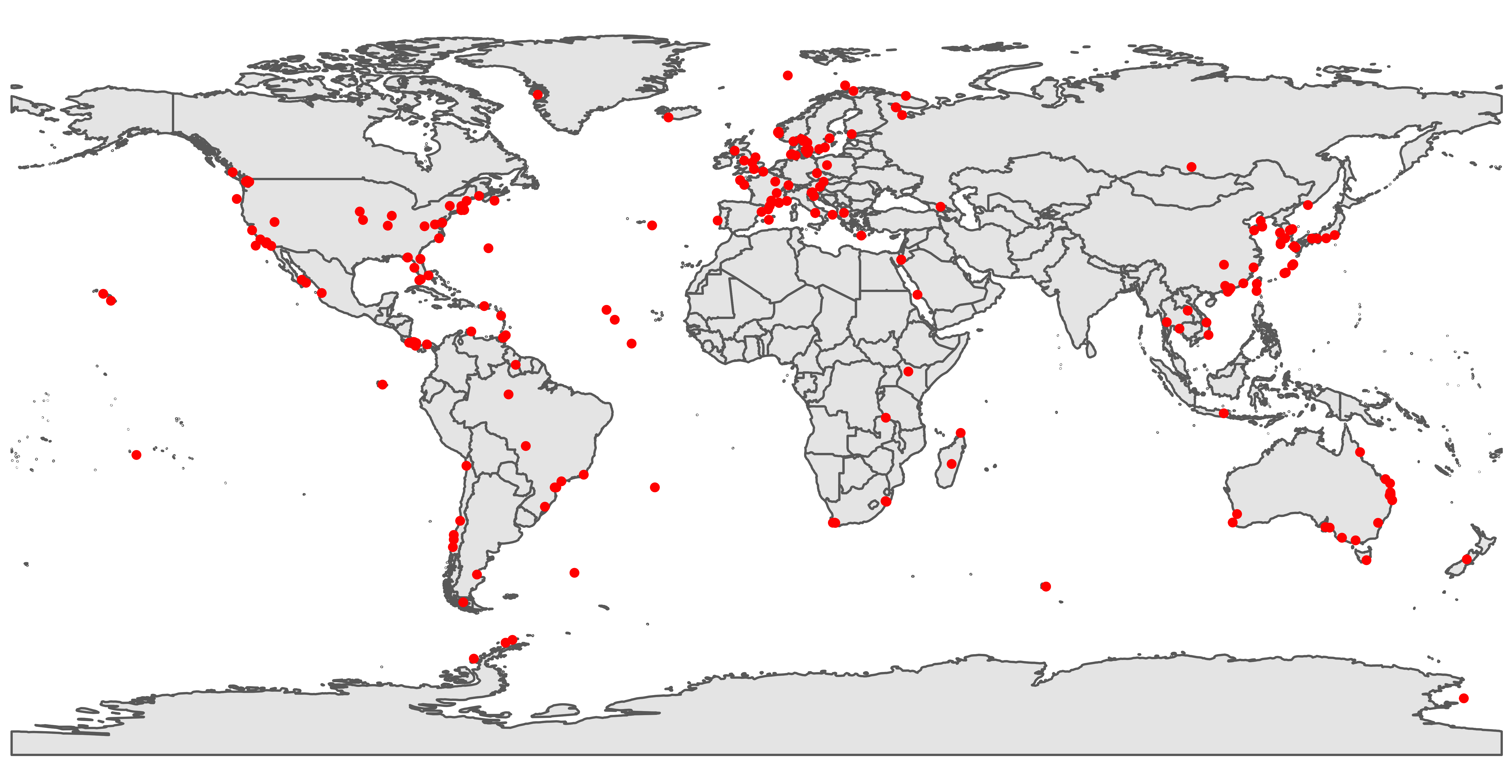
Geographical distribution of sampled accessions. See Figure 2B.
Disputed_nodes.png)
Approximate philogeny of metazoans. See Figure 2A.
Authors
Pau Alfonso (Instituto de Biología Integrativa de Sistemas, CSIC-Universitat de València), Anamarija Butković (Institut Pasteur, Université Paris Cité), Rosa Fernández (Institute of Evolutionary Biology, CSIC-Universitat Pompeu Fabra), Ana Riesgo (Museo de Ciencias Naturales, CSIC, & Department of Life Sciences, Natural History Museum of London), Santiago F. Elena (Instituto de Biología Integrativa de Sistemas, CSIC-Universitat de València)